Our researchers have developed the first comprehensive "Atlas” of regulatory areas active in gliomas
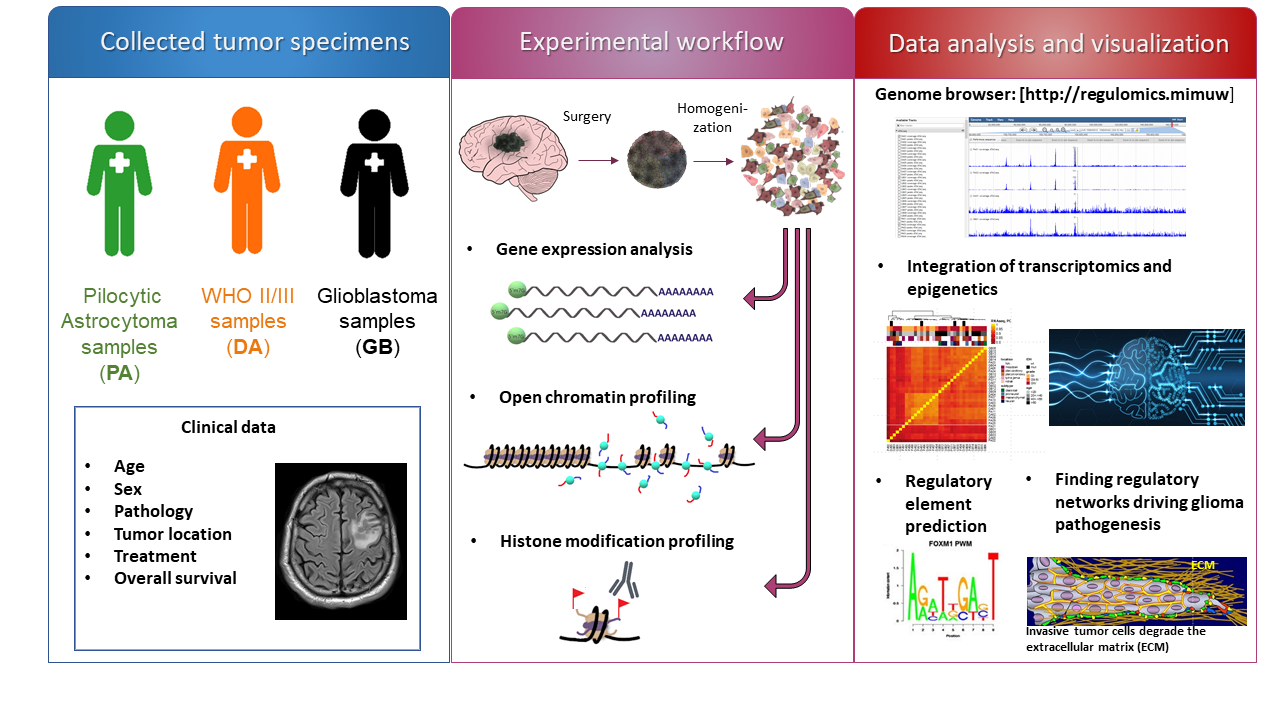
Researchers from the Nencki Institute PAS, the University of Warsaw, and the Institute of Computer Science PAS, have developed the first comprehensive "Atlas” of regulatory areas active in gliomas of various degrees of malignancy, which revealed deregulation of mechanisms controlling gene expression and a new mechanism driving the invasion of malignant brain tumors. The research was published in Nature Communications on June 15, 2021.
The interdisciplinary project Symphony 3 entitled "Atlas of regulatory areas specific for the human brain - a new tool for discovering the pathways causing selected brain diseases" has been carried out by the teams of prof. Bożena Kamińska (Nencki Institute of Experimental Biology of the Polish Academy of Sciences), dr. hab. Bartosz Wilczyński (Faculty of Mathematics, Informatics and Mechanics, University of Warsaw), and dr. Michał J Dąbrowski (Institute of Computer Science of the Polish Academy of Sciences). The project was financed by the National Research Center (NCN).
The most important findings:
- Revealing the mechanisms regulating gene expression in benign and malignant gliomas.
- Predicting a new pathway for regulating gene expression related to glioma malignancy, and confirming that this pathway controls cell migration and invasiveness in glioblastoma cells.
- Creating and sharing an atlas of active regulatory regions in gliomas of various degrees of malignancy and in the brain [http://regulomics.mimuw.edu.pl/].
The human genome is a vast set of instructions that are read and interpreted to produce cellular proteins and enable proper functions of cells and tissues. Less than 2% of the human DNA encodes proteins, therefore decoding the functions of the remaining non-coding regions is a great challenge. There are tens of thousands of genes active in each tissue, and understanding how they are regulated, would allow us to better understand the functions of the cell. In a cell, a strand of DNA is wrapped around proteins called histones, and forms a highly organized structure called chromatin. Biochemical changes in histones contribute to the openness, or lack of access to chromatin, and can stimulate or inhibit gene expression (these processes are called epigenetics). Enzymes can access and read instructions contained within DNA only at the open chromatin sites. Genome-wide mapping of regulatory regions and open chromatin provides a glimpse how genes are regulated in specific cells, and under physiological or pathological conditions. Changes in the chromatin openness are regulated by epigenetic processes that ensure their endurance, influence the reading of specific genes and, in result, drive cellular processes. Deregulation of gene expression often occurs during tumor development. The processes regulating chromatin openness are reversible, and can be adjusted externally, therefore controlling chromatin openness has a high clinical potential.
Gliomas are brain tumors, in which the proper regulation of gene expression is frequently impaired, resulting in uncontrolled tumor growth and impairment of brain functions. Malignant gliomas are most common in the elderly, are resistant to standard therapy, and have a very poor prognosis. Benign gliomas occur mainly in children, and have a better prognosis, but if left untreated, they can develop into malignant tumors.
In collaboration with neurosurgeons from Warsaw hospitals, a unique set of tumor samples was collected, and a comprehensive, whole-genome analysis of epigenetic patterns in samples of benign and malignant tumors was performed. The comparison of the patterns revealed specific processes related to malignant tumorigenesis. For the first time, researchers investigated simultaneously, the patterns of chromatin openness, histone changes, DNA methylation, and gene expression, in more than 30 brain tumor samples. All molecular clues were used to identify regulatory elements, such as promoters that control the expression of adjacent genes, and enhancers that direct the expression of distant genes.
The scientists' Atlas, which can be accessed via a web server, provides a better understanding of the importance of the non-coding regions of the genome that are active within the human brain. Moreover, findings revealed new mechanisms that control tumorigenesis in brain tumors.
“Our findings have led to the construction of the first comprehensive atlas of active regulatory elements in gliomas, which enabled the identification of functional regions such as enhancers and promoters in patient samples. This comprehensive approach revealed epigenetic patterns influencing gene expression in benign gliomas, and a novel tumor-related mechanism involving the FOXM1-driven signaling pathway controlling glioblastoma cell invasion and migration. Atlas provides a gigantic dataset that can be used for further analyses and comparisons with existing and new datasets. This will inspire new discoveries, and allow better understanding of glioblastoma development”, says Dr. Karolina Stępniak and Dr. Jakub Mieczkowski, the first authors of the publication.
Building and sharing an atlas of active regulatory regions in gliomas of various degrees of malignancy and in the brain, will enable new discoveries and a better understanding of the key mechanisms of glioblastoma development.
Karolina Stępniak, Magdalena A. Machnicka, Jakub Mieczkowski, Anna Macioszek, Bartosz Wojtaś, Bartłomiej Gielniewski, Katarzyna Poleszak, Malgorzata Perycz, Sylwia K. Król, Rafał Guzik, Michał J. Dąbrowski, Michał Dramiński, Marta Jardanowska, Ilona Grabowicz, Agata Dziedzic, Hanna Kranas, Karolina Sienkiewicz, Klev Diamanti, Katarzyna Kotulska, Wiesława Grajkowska, Marcin Roszkowski, Tomasz Czernicki, Andrzej Marchel, Jan Komorowski, Bożena Kamińska, Bartek Wilczyński. Mapping chromatin accessibility and active regulatory elements reveals pathological mechanisms in human gliomas. DOI: 10.1038/s41467-021-23922-2



